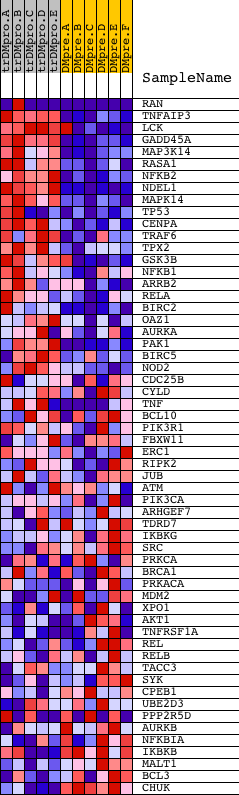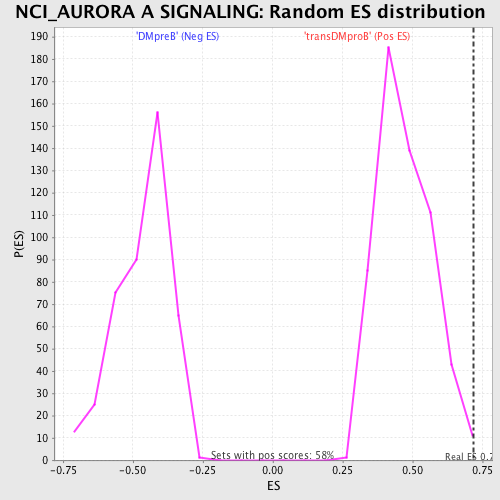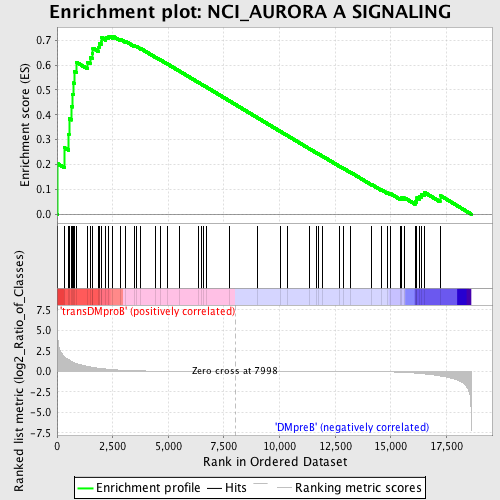
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | NCI_AURORA A SIGNALING |
| Enrichment Score (ES) | 0.7168795 |
| Normalized Enrichment Score (NES) | 1.5189409 |
| Nominal p-value | 0.0052173915 |
| FDR q-value | 0.20749149 |
| FWER p-Value | 0.956 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RAN | 5356 9691 | 22 | 4.446 | 0.2033 | Yes | ||
| 2 | TNFAIP3 | 19810 | 331 | 1.761 | 0.2677 | Yes | ||
| 3 | LCK | 15746 | 521 | 1.410 | 0.3224 | Yes | ||
| 4 | GADD45A | 17129 | 540 | 1.376 | 0.3847 | Yes | ||
| 5 | MAP3K14 | 11998 | 645 | 1.197 | 0.4342 | Yes | ||
| 6 | RASA1 | 10174 | 698 | 1.106 | 0.4823 | Yes | ||
| 7 | NFKB2 | 23810 | 744 | 1.056 | 0.5284 | Yes | ||
| 8 | NDEL1 | 20399 | 774 | 1.022 | 0.5739 | Yes | ||
| 9 | MAPK14 | 23313 | 872 | 0.931 | 0.6115 | Yes | ||
| 10 | TP53 | 20822 | 1359 | 0.593 | 0.6126 | Yes | ||
| 11 | CENPA | 8736 | 1487 | 0.534 | 0.6303 | Yes | ||
| 12 | TRAF6 | 5797 14940 | 1581 | 0.489 | 0.6478 | Yes | ||
| 13 | TPX2 | 14790 | 1607 | 0.478 | 0.6684 | Yes | ||
| 14 | GSK3B | 22761 | 1874 | 0.364 | 0.6708 | Yes | ||
| 15 | NFKB1 | 15160 | 1890 | 0.360 | 0.6866 | Yes | ||
| 16 | ARRB2 | 20806 | 1981 | 0.330 | 0.6969 | Yes | ||
| 17 | RELA | 23783 | 1985 | 0.329 | 0.7119 | Yes | ||
| 18 | BIRC2 | 4397 4398 | 2196 | 0.267 | 0.7129 | Yes | ||
| 19 | OAZ1 | 9498 | 2327 | 0.238 | 0.7169 | Yes | ||
| 20 | AURKA | 14325 | 2507 | 0.199 | 0.7164 | No | ||
| 21 | PAK1 | 9527 | 2858 | 0.127 | 0.7034 | No | ||
| 22 | BIRC5 | 8611 8610 4399 | 3080 | 0.097 | 0.6959 | No | ||
| 23 | NOD2 | 6384 | 3468 | 0.062 | 0.6779 | No | ||
| 24 | CDC25B | 14841 | 3547 | 0.056 | 0.6763 | No | ||
| 25 | CYLD | 18532 | 3768 | 0.043 | 0.6664 | No | ||
| 26 | TNF | 23004 | 4411 | 0.021 | 0.6328 | No | ||
| 27 | BCL10 | 15397 | 4654 | 0.016 | 0.6205 | No | ||
| 28 | PIK3R1 | 3170 | 4940 | 0.013 | 0.6057 | No | ||
| 29 | FBXW11 | 20926 | 5509 | 0.008 | 0.5755 | No | ||
| 30 | ERC1 | 1013 17021 995 1136 | 6373 | 0.004 | 0.5292 | No | ||
| 31 | RIPK2 | 2528 15935 | 6498 | 0.004 | 0.5227 | No | ||
| 32 | JUB | 21834 3339 | 6564 | 0.004 | 0.5193 | No | ||
| 33 | ATM | 2976 19115 | 6723 | 0.003 | 0.5110 | No | ||
| 34 | PIK3CA | 9562 | 7760 | 0.001 | 0.4552 | No | ||
| 35 | ARHGEF7 | 3823 | 8994 | -0.002 | 0.3888 | No | ||
| 36 | TDRD7 | 16217 | 10032 | -0.005 | 0.3332 | No | ||
| 37 | IKBKG | 2570 2562 4908 | 10346 | -0.006 | 0.3166 | No | ||
| 38 | SRC | 5507 | 11341 | -0.009 | 0.2635 | No | ||
| 39 | PRKCA | 20174 | 11674 | -0.011 | 0.2461 | No | ||
| 40 | BRCA1 | 20213 | 11747 | -0.011 | 0.2427 | No | ||
| 41 | PRKACA | 18549 3844 | 11943 | -0.012 | 0.2328 | No | ||
| 42 | MDM2 | 19620 3327 | 12679 | -0.018 | 0.1940 | No | ||
| 43 | XPO1 | 4172 | 12872 | -0.020 | 0.1845 | No | ||
| 44 | AKT1 | 8568 | 13176 | -0.023 | 0.1693 | No | ||
| 45 | TNFRSF1A | 1181 10206 | 14144 | -0.040 | 0.1190 | No | ||
| 46 | REL | 9716 | 14584 | -0.054 | 0.0978 | No | ||
| 47 | RELB | 17942 | 14836 | -0.066 | 0.0873 | No | ||
| 48 | TACC3 | 10021 | 14970 | -0.073 | 0.0835 | No | ||
| 49 | SYK | 21636 | 15422 | -0.112 | 0.0644 | No | ||
| 50 | CPEB1 | 8778 | 15491 | -0.119 | 0.0662 | No | ||
| 51 | UBE2D3 | 7253 | 15623 | -0.136 | 0.0654 | No | ||
| 52 | PPP2R5D | 962 1523 22957 10139 | 16091 | -0.216 | 0.0502 | No | ||
| 53 | AURKB | 20832 | 16155 | -0.226 | 0.0571 | No | ||
| 54 | NFKBIA | 21065 | 16172 | -0.230 | 0.0668 | No | ||
| 55 | IKBKB | 4907 | 16287 | -0.259 | 0.0726 | No | ||
| 56 | MALT1 | 6274 | 16378 | -0.279 | 0.0806 | No | ||
| 57 | BCL3 | 8654 | 16506 | -0.311 | 0.0881 | No | ||
| 58 | CHUK | 23665 | 17214 | -0.556 | 0.0755 | No |